pUNO1-hTMPRSS2a
-
Cat.code:
puno1-htps2a
- Documents
ABOUT
Human TMPRSS2 coding sequences
GENE DESCRIPTION
Human TMPRSS2 (transmembrane protease serine 2) is a multimeric protein containing four domains, a type II transmembrane domain, a receptor class A domain, a scavenger receptor cysteine-rich domain and a C-terminal ectodomain encompassing a large serine protease subunit. It is widely expressed in epithelial tissues, including the prostate, pancreas, liver, kidney, lung, colon, and small intestine [1]. TMPRSS2 is capable of autoactivation, and its protease domain is thought to be secreted upon autocleavage [2].
In the context of SARS-CoV-2 (2019-nCoV) infection, TMPRSS2 exerts a crucial proteolytic activation of the virus Spike protein bound to host ACE2 to facilitate the viral entry into target cells [3-6].
The hTMPRSS2 gene has two spliced transcript variants, hTMPRSS2a and hTMPRSS2b, encoding a long (1) and a short (2) isoform, respectively. The isoform 2 has an alternate 5' exon and uses a downstream AUG start codon, resulting in a shorter N-terminus as compared to the isoform 1. Both isoforms have been found to cleave the Spike protein of SARS-CoV for cathepsin L-independent entry into target cells [7]. (See Details and Specifications for more information).
 InvivoGen also offers:
InvivoGen also offers:
• SARS-CoV-2-Cellular Receptor Genes
• SARS-CoV-2-Structural Genes
PLASMIDS DESCRIPTION
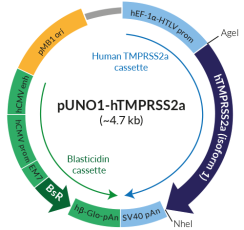
pUNO1-hTMPRSS2a and pUNO1-hTMPRSS2b feature a potent mammalian expression cassette comprised of the ubiquitous human EF1α-HTLV composite promoter and the SV40 polyadenylation (pAn) signal. Both ORFs are flanked by unique restriction sites (AgeI and NheI) to facilitate their subcloning. Both plasmids are selectable with blasticidin in E. coli and mammalian cells. They can be used for transient or stable transfection. These plasmids contain no tag.
QUALITY CONTROL
- Fully sequenced ORF
- Predominant supercoiled conformation
hTMPRSS2a is also available with hACE2 in the pDUO2-hACE2-TMPRSS2a plasmid.
If the blasticidin resistance gene does not fit your research purposes, hTMPRSS2a is also available in an expression vector selectable with hygromycin (upon request).
![]() Read our reviews about SARS-CoV-2.
Read our reviews about SARS-CoV-2.
References
1. Bugge T.H. et al., 2009. Type II transmembrane serine proteases. J. Biol. Chem. 284(35) :23177-23181.
2. Afar D.E.H, et al., 2001. Catalytic Cleavage of the Androgen-regulated TMPRSS2 Protease Results in Its Secretion by Prostate and Prostate Cancer Epithelia. Cancer Res. 61(4):1686-1692.
3. Li W. et al., 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 426(6965):450-454.
4. Hoffmann M. et al., 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 181:1-16.
5. Zhou P. et al., 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 579(7798):270-273.
6. Hoffmann M. et al., 2020. A multibasic cleavage site in the Spike protein of SARS-CoV-2 is essential for infection of human lung cells. Molecular Cell. 78:1-6.
7. Zmora P. et al., 2015. TMPRSS2 isoform 1 activates respiratory viruses and is expressed in viral target cells. PLoS ONE 10(9):e0138380.
All products are for internal research use only, and not for human or veterinary use.
SPECIFICATIONS
Specifications
Plasmid construct has been confirmed by restriction analysis and full-length open reading frame (ORF) sequencing.
CONTENTS
Contents
-
Product:pUNO1-hTMPRSS2a
-
Cat code:puno1-htps2a
-
Quantity:20 µg
2 x 1 ml blasticidin at 10 mg/ml
Shipping & Storage
- Shipping method: Room temperature
- -20°C
- Avoid repeated freeze-thaw cycles
Storage:
Caution:
Details
Human TMPRSS2 (transmembrane protease serine 2) is a type II transmembrane serine protease (TTSP) protein containing four subunits: an N-terminal transmembrane domain, and a large ectodomain comprised of an LDLA (low-density lipoprotein receptor A) subunit, a group A scavenger receptor domain, and a trypsin-like serine protease domain in C-terminal [1]. TTSPs activation requires cleavage following a basic amino acid residue (Arg for TMPRSS2) in a conserved activation motif preceding the catalytic domain [1].
TMPRSS2 has been found to cleave the hemagglutinin (HA) glycoprotein from the influenza virus, and the Spike glycoprotein of MERS-CoV, SARS-CoV, and SARS-CoV-2, to generate unlocked, fusion-catalyzing HA and Spike at the target cell surface [2-4].
In the context of SARS-CoV-2 infection, TMPRSS2 exerts a crucial proteolytic activation of the S protein bound to ACE2 to facilitate the viral entry into target cells [2, 6, 7]. The Spike glycoprotein harbors two distinct cleavage sites, known as S1/S2 and S2’, at the intersection its S1 and S2 domains, and inside the S2 domain, respectively. While the exact timing and location for Spike binding, cleavage, and fusion processes to take place remain to be determined, it has been proposed that the S protein is cleaved into two subunits (S1 and S2) by proteases, including furin and TMPRSS2 [2, 8, 9]. S1 binds to ACE2, and S2 is further cleaved and activated by the TMPRSS2 [2, 8]. Together these actions result in host-viral membrane fusion and the viral RNA genome is released into the host cell cytoplasm.
Interestingly, while TTSPs are believed to remain membrane-associated after activation because of a disulfide bond linking the pro-domain and the catalytic domain [1], TMPRSS2 has been found to be autoactivated, cleaved and its serine protease domain released in the tissues, cell line cultures, and mouse serum [10].
Two spliced transcript variants encoding a long (1) and a short (2) isoform have been found for the TMPRSS2 gene, and both isoforms have been found to cleave the Spike protein of SARS-CoV for cathepsin L-independent entry into target cells [3].
Camostat and Nafamostat are two clinically-proven inhibitors of TMPRSS2 [1, 11].
References
1. Bugge T.H. et al. 2009. Type II transmembrane serine proteases. J. Biol. Chem. 284(35) :23177-23181.
2. Hoffmann M. et al. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 181:1-16.
3. Zmora P. et al. 2015. TMPRSS2 isoform 1 activates respiratory viruses and is expressed in viral target cells. PLoS ONE 10(9):e0138380.
4. Matsuyama S. et al. 2010. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 84:12658-12664.
5. Hamming I. et al. 2004. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. J. Pathol. 203:631-637.
6. Li W. et al. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 426(6965):450-454.
7. Zhou P. et al. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 579(7798):270-273.
8. Walls A.C., et al. 2020. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 181(2):281-292.e6.
9. Hoffmann M. et al., 2020. A multibasic cleavage site in the Spike protein of SARS-CoV-2 is essential for infection of human lung cells. Molecular Cell. 78:1-6.
10. Afar, D.E.H. et al. 2001. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res. 61(4):1686-1692.
11. Vincent, M.J. et al. 2005. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J 2, 69.
DOCUMENTS
Documents
Technical Data Sheet
Plasmid Map and Sequence
Plasmid Sequence
Safety Data Sheet
Certificate of analysis
Need a CoA ?