293-SARS2-S Cells
-
Cat.code:
293-cov2-s
- Documents
ABOUT
SARS-CoV-2 Spike (D614)-expressing 293 cells
InvivoGen offers human embryonic kidney (HEK)-293-derived cells, specifically designed for COVID-19 studies:
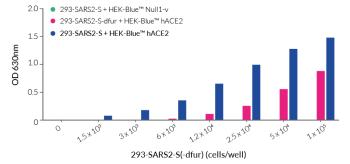
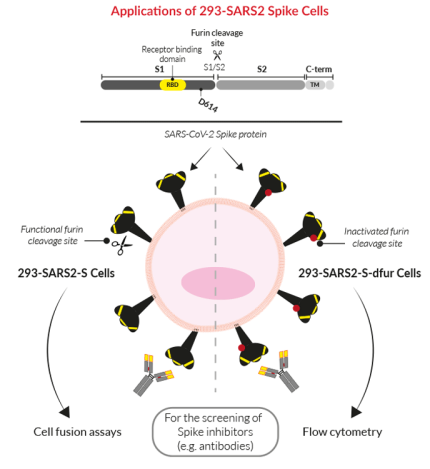
• 293-SARS2-S Cells: with a functional furin cleavage site and recommended for cell-cell fusion assays with ACE2-expressing cells.
• 293-SARS2-S-dfur Cells: with an inactive furin (dfur) cleavage site facilitating improved surface expression and detection for flow cytometry.
They have been engineered to overexpress the original Wuhan‑Hu-1 SARS‑CoV-2 Spike (S) gene, and for optimal expression, the C‑terminal ER-retention signal has been removed [1,2]. Importantly, the Spike protein contains a furin cleavage site that can affect its cellular expression [3].
A crucial step in developing successful COVID-19 treatments is the validation of their efficacy using rapid, reliable and robust platforms. To this end, cell-based assays using Spike-expressing cells provide a more relevant in vivo biological context than biochemical assays (e.g. ELISAs).
Key Features
- Stable expression of the original SARS-CoV-2 Spike (D614) protein.
- Functionally validated in cell fusion assays with HEK-Blue™ hACE2(-TMPRSS2) Cells (293-SARS2-S Cells) and by flow cytometry (293-SARS2-S-dfur Cells).
Applications
- 293-SARS2-S-dfur Cells: Screening patient/vaccinated sera and/or monoclonal antibodies against the SARS-CoV-2 Spike by flow cytometry.
- 293-SARS2-S Cells: Cell fusion assays for screening novel inhibitors of the SARS-CoV-2 Spike-host interaction using InvivoGen's permissive reporter cell lines (HEK-Blue™ hACE2(-TMPRSS2) and A549-Dual™ hACE2-TMPRSS2).
![]() Learn more about InvivoGen's tools for cell fusion assays
Learn more about InvivoGen's tools for cell fusion assays
➔ 293-SARS2-S-dfur cells have also been generated with the Spike of other variants of concern: Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (BA.1).
These cells have been used to compare the activity of anti-SARS-CoV-2 Spike antibodies. See data
References:
1. Johnson, M.C. et al. 2020. Optimized Pseudotyping Conditions for the SARS-COV-2 Spike Glycoprotein. J Virol 94.
2. Ou, X. et al. 2020. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 11, 1620.
3. Coutard, B. et al. 2020. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res 176, 104742.
Disclaimer: These cells are for internal research use only and are covered by a Limited Use License (See Terms and Conditions). Additional rights may be available.
Available upon request
- 293-SARS2-S-V2-dfur cells (Alpha/B.1.1.7/U.K variant)
- 293-SARS2-S-V3-dfur cells (Beta/B.1.351/S.A variant)
- 293-SARS2-S-V5-dfur cells (Gamma/P.1/BRA variant)
- 293-SARS2-S-V8-dfur cells (Delta/B.1.617.2/IND variant)
- 293-SARS2-S-V11-dfur cells (Omicron/BA.1 variant)
SPECIFICATIONS
Specifications
Cell fusion assays, Flow cytometry
Complete DMEM (see TDS)
Verified using Plasmotest™
Each lot is functionally tested and validated.
CONTENTS
Contents
-
Product:293-SARS2-S Cells
-
Cat code:293-cov2-s
-
Quantity:3-7 x 10^6 cells
- 1 ml of Blasticidin (10 mg/ml)
- 1 ml of Normocin™ (50 mg/ml). Normocin™ is a formulation of three antibiotics active against mycoplasmas, bacteria, and fungi.
Each cell line is sold separately. See TDS for the exact contents of each cell line.
Shipping & Storage
- Shipping method: Dry ice
- Liquid nitrogen vapor
Storage:
Details
SARS-CoV-2 Spike
Spike (S) is a structural glycoprotein expressed on the surface of SARS‑CoV-2. It mediates membrane fusion and viral entry into target cells upon binding to the host receptor, ACE2 and host proteolytic cleavage (e.g. TMPRSS2 and furin). The S protein consists of an N-terminal ectodomain, a transmembrane anchor, and a short C-terminal cytoplasmic tail. The ectodomain contains the S1 subunit, which encodes the receptor-binding domain (RBD), a key target in treatment and vaccination strategies against COVID-19, as well as the S2 subunit, needed for membrane fusion. Notably, the C-terminal cytoplasmic tail of the S protein encodes a presumptive endoplasmic reticulum (ER)-retention motif (KxHxx), which has previously been shown to enable the accumulation of SARS-CoV S proteins at the ER-Golgi intermediate compartment (ERGIC) and facilitate their incorporation into new virions.
DOCUMENTS
Documents
Technical Data Sheet
Validation Data Sheet
Safety Data Sheet
Certificate of analysis
Need a CoA ?