Pseudoparticle infection and cell-fusion assays
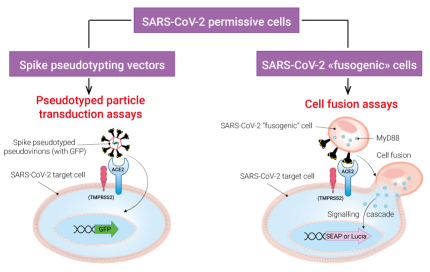
InvivoGen offers a collection of biological tools to assess two key functions of the SARS-CoV-2 Spike protein: infection of target cells and cell fusion. Depending on your needs, you may choose among families of:
• SARS-CoV-2 permissive (target) cells
• Spike pseudotyping vectors
• SARS-CoV-2 "fusogenic" cells
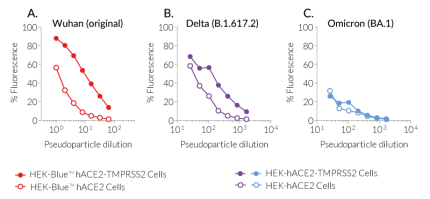
Below are the data obtained with target cells exposed to Spike from the Wuhan original strain as well as from two variants of concern (VOCs), Delta and Omicron, which have been reported to differ in their infection and pathogenic capabilities.
SARS-CoV-2 permissive (target) cells:
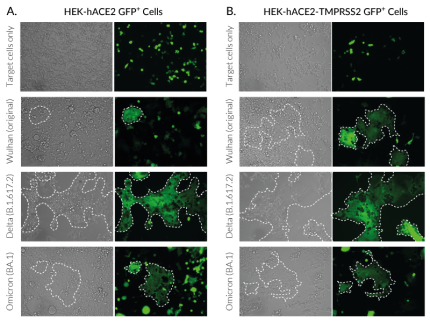
InvivoGen has developed a series of target cells expressing high levels of the host receptor ACE2 alone, or in combination with the host protease TMPRSS2 at their cell surface. Thus, these cells are permissive to infection by SARS-CoV-2 and Spike-pseudotyped lentiviral particles (Figure 1). Moreover, they allow cell fusion and formation of syncytia (Figure 2), illustrating one major SARS-COV-2 cytopathic effect.
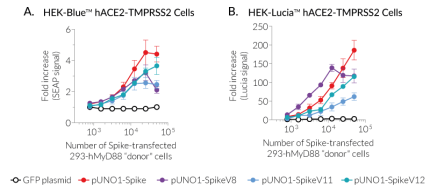
SARS-CoV-2 permissive cells express NF-κB- and IRF-inducible reporter genes encoding the Lucia™ luciferase or the secreted embryonic alkaline phosphatase (SEAP) to assess innate sensing pathways, or Spike-mediated fusogenicity using InvivoGen's cell fusion assays (Figure 3). These cells are also available without reporter genes.
HEK293-derived cell lines:
- HEK-Blue™ hACE2(-TMPRSS2) cells express an NF-κB-inducible SEAP reporter gene.
- HEK-Lucia™ hACE2(-TMPRSS2) cells express an NF-κB-inducible Lucia luciferase reporter gene. Available upon request.
- HEK-hACE2(-TMPRSS2) cells do not express any inducible reporter gene. Available upon request.
A5493-derived cell lines:
- A549-hACE2-(TMPRSS2) cells do not express any inducible reporter gene.
- A549-Dual™ (KO-MDA5 or KO-RIG-I) hACE2-TMPRSS2 cells express two inducible reporter genes, allowing the concomitant study of the IRF and NF-κB pathways by monitoring the Lucia™ luciferase and SEAP, respectively.
Spike pseudotyping vectors for lentiviral assays:
InvivoGen offers a collection of pLV-Spike plasmids specifically designed for pseudotyping lentiviral particles with the full-length SARS-CoV-2 Spike protein from various variants. This family encompasses the first identified SARS-CoV-2 (Wuhan) and major variants that rapidly arose during the pandemic progression. Spike-mediated cell infection has been tested in-house using pLV-Spike plasmids for the Wuhan, Delta and Omicron variants together with ACE2(-TMPRSS2)-expressing HEK293-derived cells as permissive cells (Figure 1).
- pLV-Spike (D614) encodes the Spike protein from the Wuhan (original) variant.
- pLV-SpikeV8 encodes the Spike protein from the Delta (B.1.617.2) variant.
- pLV-SpikeV11 encodes the Spike protein from the Omicron (BA.1) variant.
Browse the whole collection of pLV-Spike plasmids.
SARS-COV-2 Spike "fusogenic" cells:
InvivoGen has developed a simple cell fusion assay to assess SARS-CoV-2 Spike-mediated fusogenicity. Briefly, fusogenic ("donor") cells expressing Spike at the cell surface and a constitutively active human MyD88 adaptor molecule in the cytosol are co-cultured with permissive SARS-CoV-2 target ("acceptor") cells expressing an NF-kB-SEAP or -Lucia™ luciferase inducible reporter gene. Upon Spike-ACE2-dependent cell fusion, the active MyD88 adaptor is transferred from the "donor" to the "acceptor" cells allowing the reporter gene expression.
Depending on your needs, you may choose from "fusogenic" cells either transiently or stably transfected with a gene encoding the SARS-CoV-2 Spike protein:
- To compare the fusogenicity of SARS-CoV-2 variants, Spike-mediated cell fusion can be tested using MyD88-expressing HEK293 cells transiently transfected with Spike plasmids and HEK-Blue™ or -Lucia™ hACE2(-TMPRSS2) cells (Figure 3).
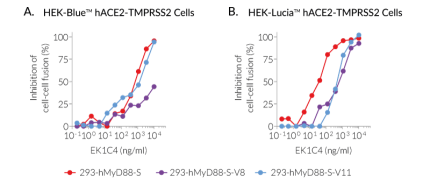
- To screen for antibodies or small molecules inhibiting cell fusion, MyD88-expressing HEK293 cells with stable Spike expression can be used as these have been selected on their fusogenic potency. "Donor" cells with stable expression of the Wuhan (original), Delta, or Omicron Spike have been functionally tested in a cell-fusion inhibition assay using the small molecule EK1C4 (Figure 4).
MyD88-expressing HEK293 cells:
- 293-hMyD88 cells express a constitutively active human MyD88 adaptor molecule.
Spike plasmids for cellular expression:
- pUNO1-Spike encodes the Spike protein from the Wuhan (original) variant.
- pUNO1-SpikeV8 encodes the Spike protein from the Delta (B.1.617.2) variant.
- pUNO1-SpikeV11 encodes the Spike protein from the Omicron (BA.1) variant.
- pUNO1-SpikeV12 encodes the Spike protein from the Omicron (BA.2) variant.
Browse the whole collection of pUNO1-Spike plasmids.
MyD88-expressing HEK293 cells with stable Spike expression:
- 293-hMyD88-S cells stably express Spike from the Wuhan (original) strain and human MyD88. Available upon request.
- 293-hMyD88-S-V8 cells stably express Spike from the Delta (B.1.617.2) variant and human MyD88. Available upon request.
- 293-hMyD88-S-V11 cells stably express Spike from the Omicron (BA.1) variant and human MyD88. Available upon request.